EGFR endocytosis in signaling and cancer
Endocytosis is becoming increasingly recognized as a central component of the cell signaling machinery, acting not only as a platform for the integration of multiple signaling events, but also as a mechanism for achieving spatial and temporal regulation of signaling [1,2]. Not surprisingly, endocytosis is implicated in the regulation of many cellular processes and is increasingly being linked to diseases, such as cancer. A long-standing goal of our lab is to understand how endocytosis regulates cell signaling and how its deregulation might contribute to cancer. We are using the epidermal growth factor receptor (EGFR), a known oncogene, as a “model endocytic system” to characterize novel endocytic mechanisms and their contribution to tumorigenesis.
Our work has highlighted how the choice of endocytic route is decisive in determining receptor fate and signaling outcomes. Depending on ligand concentration, EGFR internalization can occur via the well-characterized clathrin-mediated endocytosis (CME) route or the alternative non-clathrin mediated endocytosis (NCE) route, recently identified in our lab [3-6]. While CME is involved in receptor recycling and maintaining signaling, NCE targets the EGFR to degradation in the presence of high ligand concentrations, causing signal extinction. Thus, NCE could potentially act as a tumor suppressor pathway preventing overstimulation of the EGFR pathway, while CME, by maintaining EGFR signaling could act as an oncogenic mechanism.
To understand better the role of endocytosis in signal regulation and cancer, we have performed an in-depth characterization of the NCE pathway using a proteomics approach and advanced imaging and cellular biology techniques. NCE represents a novel endocytic mechanism, in which the endoplasmic reticulum (ER) makes physical contacts with the plasma membrane (PM) to allow EGFR internalization and targeting to lysosomal degradation [6]. We are now investigating the functional relevance of NCE in physiological processes (e.g., proliferation, migration, mitochondrial function, and cell metabolism), and its alterations in cancer. We are also employing systems biology approaches to understand how NCE and CME are integrated and to define the parameters that trigger the switch between the two endocytic pathways [5]. Finally, thanks to the ‘Novel DiagnosticsProgram’ at IEO, we have access to the necessary resources for preclinical research into the pathological relevance of EGFR endocytic pathways in cancer.
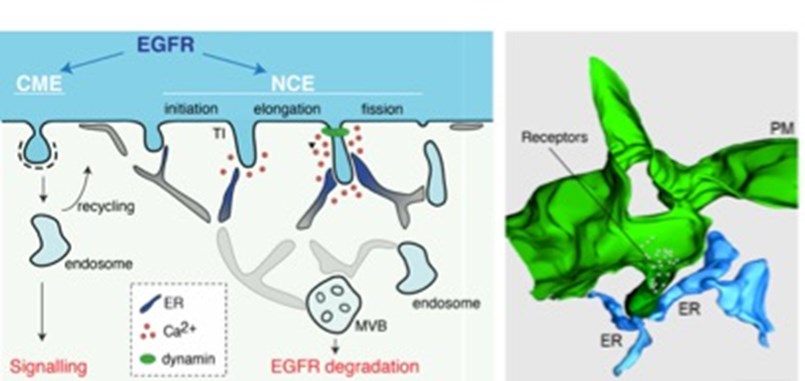
Figure 1. Left, Schematic representation of the different EGFR endocytic routes.The mechanism for NCE vesicle maturation involving ER:PM contacts and local Ca2+release is shown. Right, 3D reconstruction of NCE tubular invaginations at the PM (green) in contact with the ER (light blue). Taken from [6].
Endocytosis, cancer stem cells and metastasis
A second line of research in the lab is focused on investigating the functional involvement of endocytic proteins in cancer. Through high-throughput screenings and high-resolution studies, we have identified several endocytic proteins, or their regulators (e.g. kinases, E3 ligases, deubiquitinases), that are aberrantly expressed in cancer [7-10].
One such protein is the cell fate determinant and endocytic protein, Numb, whose expression is frequently downregulated in a range of cancers, including breast and lung cancer [10,11]. Our studies have revealed that Numb, via its regulation of the tumor suppressor, p53, and the oncogene, Notch, represents a key tumor suppressor protein, which when lost promotes aggressive cancer phenotypes and associates with poor prognosis in patients [12]. At the mechanistic level, loss of Numb disrupts homeostasis of the stem cell compartment in the mammary gland by altering stem cell self-renewal kinetics and progenitor maturation, leading to the emergence of cancer stem cells [13]. In pre-clinical studies, we have shown that targeting Numb loss in breast cancer has a potentially huge therapeutic significance, since we were able to selectively eradicate the cancer stem cell population [14]. Ongoing studies performed in collaboration with the ‘Novel DiagnosticsProgram’ at IEOare aimed at exploiting this knowledge to develop novel prognostic-predictive tumor markers and innovative treatments for aggressive cancers.
Other proteins-of-interest identified in our screenings include the endocytic adaptor protein, Epsin3 [15-17], and the serine-threonine kinase, CDK12 [9]. Although little is known about the biological functions of these protein, our screenings have highlighted an association with breast cancer progression. By combining retrospective analyses of breast cancer cohorts with high-resolution in vitro andin vivo functional studies, we are investigating the role of Epsin3 and CDK12 as a prognostic marker and as a determinant in breast cancer tumorigenesis and disease progression, focusing, in particular, on their function in stem cells and in metastasis. Knowledge on the molecular mechanisms underlying Epsin3 and CDK12 function will be exploited to identify novel molecular targets for the development of new therapies for breast cancer.
Cancer-specific profiles
Using cutting-edge whole-genome technologies (coding and non-coding RNA profiling, next generation sequencing, single cell analysis, proteomics), we have derived several cancer-specific profiles, which have driven numerous high-resolution studies in our lab focused on markers that have potential diagnostic, prognostic and/or therapeutic relevance [15-25]. These include studies focused on a 10-gene signature able to predict prognosis of patients with stage I lung adenocarcinoma [19-21], and on a circulating miRNA signature capable of detecting early stage lung cancer in asymptomatic patients [22-24]. We have also obtained the transcriptional profile of purified normal human mammary SCs, which led to the identification of a mammary SC signature that is able to distinguish breast cancers based on their degree of ‘stemness” [25]. Our work has shown that aggressive, poor prognosis breast cancers display a “SC-like” transcriptional profile, while less-aggressive, good prognosis breast cancers display a“non-SC-like” profile, indicating that our signature could be useful in the prediction of prognosis in breast cancer patients. Additionally, functional studies on the individual markers that comprise the mammary SC signature are being performed with a view to uncovering mechanisms and molecular targets that could lead to the development of novel therapies for breast cancer. Knowledge from these discoveries in our basic research group are being transferred to the Novel Diagnostics Program at IEO, to promote their translation into real clinical applications for diagnostic, prognostic and therapeutic purposes.
References
- Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, Di Fiore PP. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. 2012 Physiol Rev. 92:273-366.
- Barbieri E, Di Fiore PP, Sigismund S. Endocytic control of signaling at the plasma membrane. 2016 Curr Opin Cell Biol. 39:21-27.
- Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. 2008 Dev Cell. 15:209-19.
- Sigismund S, Algisi V, Nappo G, Conte A, Pascolutti R, Cuomo A, Bonaldi T, Argenzio E, Verhoef LG, Maspero E, Bianchi F, Capuani F, Ciliberto A, Polo S, Di Fiore PP. Threshold-controlled ubiquitination of the EGFR directs receptor fate. 2013 EMBO J. 32:2140-57.
- Capuani F, Conte A, Argenzio E, Marchetti L, Priami C, Polo S, Di Fiore PP, Sigismund S, Ciliberto A. Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells. 2015 Nat Commun. 6:7999.
- Caldieri G, Barbieri E, Nappo G, Raimondi A, Bonora M, Conte A, Verhoef LGGC, Confalonieri S, Malabarba MG, Bianchi F, Cuomo A, Bonaldi T, Martini E, Mazza D, Pinton P, Tacchetti C, Polo S, Di Fiore PP, Sigismund S. Reticulon 3-dependent ER-PM contact sites control EGFR non-clathrin endocytosis. 2017 Science 356:617-24.
- Confalonieri S, Quarto M, Goisis G, Nuciforo P, Donzelli M, Jodice G, Pelosi G, Viale G, Pece S, Di Fiore PP. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. 2009 Oncogene 28:2959-68.
- Luise, C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, Viale G, Di Fiore, PP, Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. 2011 PLoS One 6:e15891.
- Capra M, Nuciforo PG, Confalonieri S, Quarto M, Bianchi M, Nebuloni M, Boldorini R, Pallotti F, Viale G, Gishizky ML, Draetta GF, Di Fiore PP. Frequent alterations in the expression of serine/threonine kinases in human cancers. 2006 Cancer Res. 66:8147-54.
- Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida, S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. 2004 J Cell Biol. 167:215-21.
- Westhoff B, Colaluca IN, D'Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, Pece S, Di Fiore PP. Alterations of the Notch pathway in lung cancer. 2009 Proc Natl Acad Sci U S A. 106:22293-98.
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP. NUMB controls p53 tumour suppressor activity. 2008 Nature451:76-80.
- Tosoni D, Zecchini S, Coazzoli M, Colaluca I, Mazzarol G, Rubio A, Caccia M, Villa E, Zilian O, Di Fiore PP, Pece S. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. 2015 J Cell Biol. 211:845-62.
- Tosoni D, Pambianco S, Ekalle Soppo B, Zecchini S, Bertalot G, Pruneri G, Viale G, Di Fiore PP, Pece S. Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers. 2017 EMBO Mol Med. 9:655-71.
- Vecchi M, Nuciforo P, Romagnoli S, Confalonieri S, Pellegrini C, Serio G, Quarto M, Capra M, Roviaro GC, Contessini Avesani E, Corsi C, Coggi G, Di Fiore PP, Bosari S. Gene expression analysis of early and advanced gastric cancers. 2007 Oncogene 26:4284-94.
- Vecchi M, Confalonieri S, Nuciforo P, Vigano MA, Capra M, Bianchi M, Nicosia D, Bianchi F, Galimberti V, Viale G, Palermo G, Riccardi A, Campanini R, Daidone MG, Pierotti MA, Pece S, Di Fiore PP. Breast cancer metastases are molecularly distinct from their primary tumors. 2008 Oncogene 27:2148-58.
- Ko G, Paradise S, Chen H, Graham M, Vecchi M, Bianchi F, Cremona O, Di Fiore PP, De Camilli P. Selective high-level expression of epsin 3 in gastric parietal cells, where it is localized at endocytic sites of apical canaliculi. 2010 Proc Natl Acad Sci USA. 107:21511-16.
- Nicassio F, Bianchi F, Capra M, Vecchi M, Confalonieri S, Bianchi M, Pajalunga D, Crescenzi M, Bonapace IM, Di Fiore PP. A cancer-specific transcriptional signature in human neoplasia. 2005 J Clin Invest. 115:3015-25.
- Bianchi F, Nuciforo P, Vecchi M, Bernard L, Tizzoni L, Marchetti A, Buttitta F, Felicioni L, Nicassio F, Di Fiore PP. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. 2007 J Clin Invest. 117:3436-44.
- Bianchi F, Nicassio F, Di Fiore PP. Unbiased vs. biased approaches to the identification of cancer signatures: the case of lung cancer. 2008 Cell Cycle 7:729-34.
- Dama, E, Melocchi, V, Dezi, F, Pirroni, S, Carletti, RM, Brambilla, D, Bertalot, G, Casiraghi, M, Maisonneuve, P, Barberis, M, Viale, G, Vecchi, M, Spaggiari, L, Bianchi, F, Di Fiore, PP. An Aggressive Subtype of Stage I Lung Adenocarcinoma with Molecular and Prognostic Characteristics Typical of Advanced Lung Cancers. 2017 Clin Cancer Res. 23:62-72.
- Bianchi F, Nicassio F, Marzi M, Belloni E, Dall'olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G, Di Fiore PP. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. 2011 EMBO Mol Med. 3:495-503.
- Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, Bertolotti R, Bellomi M, Rampinelli C, Maisonneuve P, Spaggiari L, Veronesi G, Nicassio F, Di Fiore PP, Bianchi F. miR-Test: a blood test for lung cancer early detection. 2015 JNCI 107(6):djv063.
- Marzi MJ, Montani F, Carletti RM, Dezi F, Dama E, Bonizzi G, Sandri MT, Rampinelli C, Bellomi M, Maisonneuve P, Spaggiari L, Veronesi G, Bianchi F, Di Fiore PP, Nicassio F. Optimization and Standardization of Circulating MicroRNA Detection for Clinical Application: The miR-Test Case. 2016 Clin Chem. 62:743-54.
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. 2010 Cell 140:62-73.